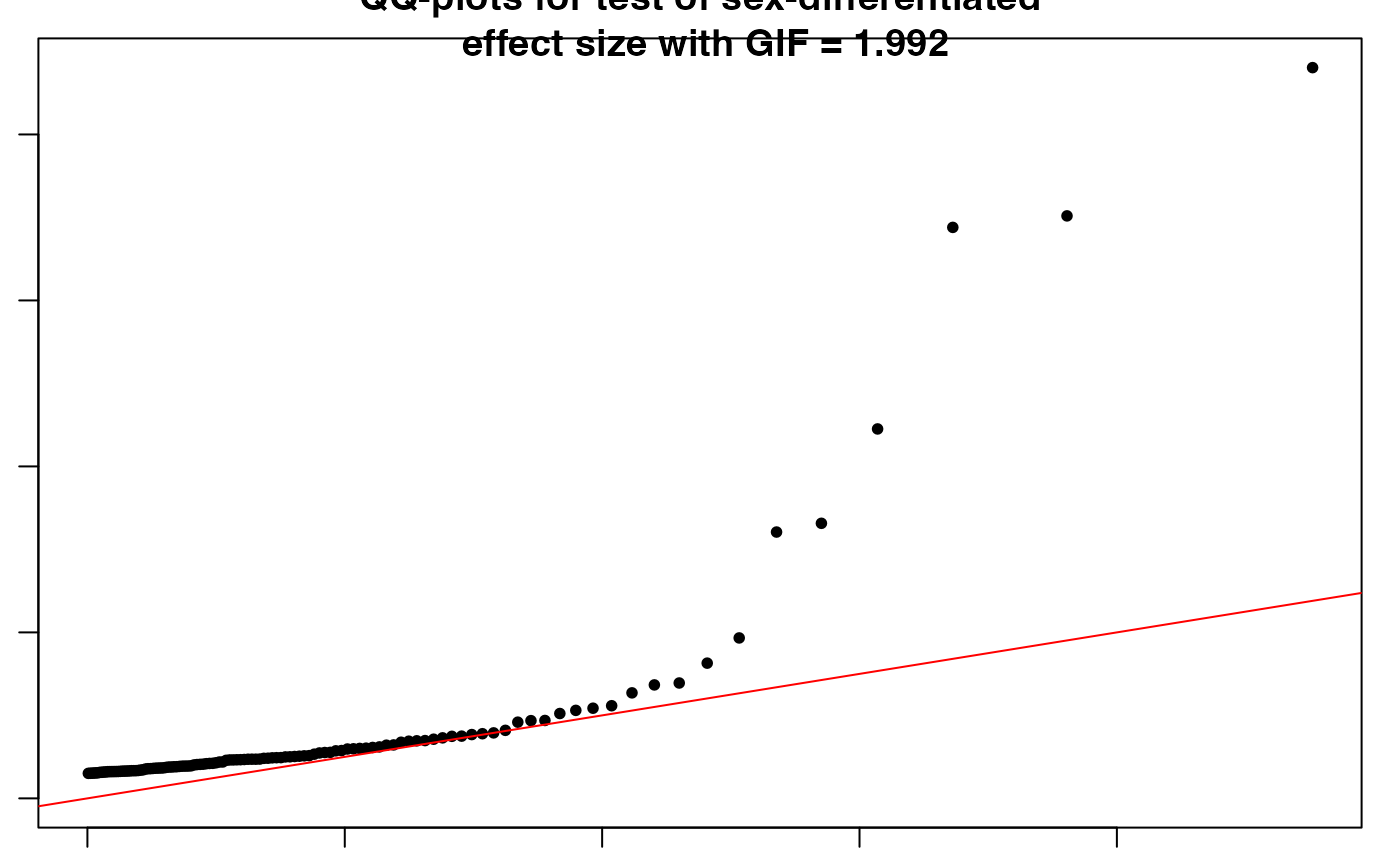
SexDiff: Sex difference in effect size for each SNP using t-test.
Source:R/GXwasR_main_functions.R
SexDiff.RdThis function uses the GWAS summary statistics from sex-stratified tests like "FMstratified", to evaluate the difference in effect size between males and females at each SNP using a t-test.
The input dataframes should only include X-chromosome in order to obtain results for sex differences based solely on X-linked loci.
Arguments
- Mfile
R dataframe of summary statistics of GWAS or XWAS of male samples with six mandatory columns, SNP(Variant),CHR(Chromosome number), BP(Base pair position),A1(Minor allele),BETA_M(Effect size) and SE_M(Standard error). This can be generated by running FM01comb or "FMstratified" model with GXWAS function.
- Ffile
R dataframe of summary statistics of GWAS or XWAS of male samples with six mandatory columns, SNP(Variant),CHR(Chromosome number), BP(Base pair position),A1(Minor allele),BETA_F(Effect size) and SE_F(Standard error). This can be generated by running FM01comb or "FMstratified" model with GXWAS function.